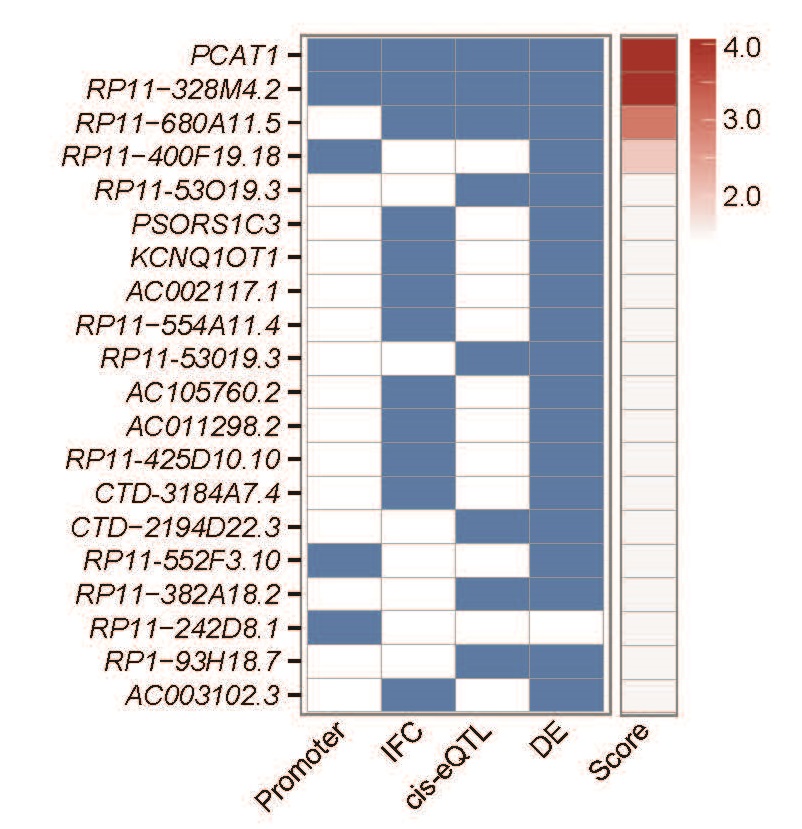
Top 20 IncRNAs associated with risk for prostate cancer. (Image supplied)
When it comes to treating prostate cancer, the news is predominantly positive: a combination of early detection and treatment by radiotherapy and surgery mean that most patients will not die of the disease. Still, its incidence continues to rise and prostate cancer remains the most frequently diagnosed non-skin malignancy in Canadian men.
A large number of patients are likely being over-treated today, a dilemma that a Nature Genetics study http://www.nature.com/ng/journal/v48/n10/full/ng.3637.html (August 2016) involving TFRI-funded researchers aimed to resolve by understanding how differences in the germline influence the type of prostate cancer that arises. The study results provide key insight into how a patient’s normal, healthy genome can change the way prostate cancer will develop, and suggests that existing diagnostics assays can be improved considering that information.
Led by TFRI New Investigator Dr. Housheng Hansen He (UHN), the international team identified 45 candidate lncRNAs associated with risk for prostate cancer by using integrative analysis of the lncRNA transcriptome with genomic data and single nucleotide polymorphisms (SNPs) data from prostate cancer genome-wide association studies (GWAS).
The mechanism underlying the top lncRNA hit, PCAT1, was then evaluated, and it was found that PCAT1 promotes prostate cancer cell proliferation and tumour growth in vitro and in vivo. A germline SNP that increases the risk of contracting cancer was identified, and demonstrated that it does so by changing the regulation of the relatively poorly understood gene. The study also identified a previously unknown function of PCAT1 in the androgen signaling pathway and that PCAT1 may promote prostate tumorigenesis through multiple pathways.
These findings suggest that modulating lncRNA expression is an important mechanism for risk-associated SNPs in promoting prostate transformation. In the future, patients who have lncRNAs, such as high-risk PCAT1, can be treated more robustly, while those who are classified as lower risk based on their genome can avoid the risks and consequences of overtreatment.
These results were validated in primary patient tumours studied in the Canadian Prostate Cancer Genome Network, a national project studying the genomics of prostate cancer. Given the success of the present study, the team suggests similar work should be performed to characterize risk-associated lncRNAs in all cancer types.
Study: Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer
Authors: Haiyang Guo, Musaddeque Ahmed, Fan Zhang, Cindy Q Yao, SiDe Li, Yi Liang, Junjie Hua, Fraser Soares, Yifei Sun , Jens Langstein , Yuchen Li , Christine Poon , Swneke D Bailey , Kinjal Desai, Teng Fei, Qiyuan Li, Dorota H Sendorek , Michael Fraser , John R Prensner, Trevor J Pugh , Mark Pomerantz, Robert G Bristow, Mathieu Lupien , Felix Y Feng , Paul C Boutros, Matthew L Freedman, Martin J Walsh & Housheng Hansen He.
Funding: P.C.B. is supported by a Terry Fox Research Institute New Investigator Award and a CIHR New Investigator Award.
Links#2