.jpg)
Top photo: Dr. Martin Koebel, study first author.
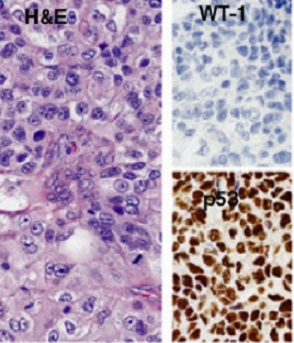
Above: Endometriod carcinoma (Grade 3) reclassified from high-grade serous carcinoma on the basis of a biomarker-assisted review (the absence of WT1) and the morphology showing a solid neoplasm (upper right) with squamoid features (lower left).
A biomarker study by COEUR (Canadian Ovarian Experimental Unified Resource), TFRI’s pan-Canadian ovarian cancer research team, examining over 2,000 ovarian cancer tissue samples has led to the development of an inexpensive immunohistochemical algorithm for classifying the five different disease subtypes. Since 2009, the COEUR team has collected the samples from patients across the country with TFRI funding. Biomarkers require large-scale validation, and the COEUR provides an ideal platform.
Ovarian cancer is the fifth-leading cause of cancer-related deaths in the western world. Over the last decade, it has become clear that it is not a single disease, but consists of five different diseases currently defined by five histotypes, each with unique molecular characteristics, risk factors and clinical behaviour. The five main histotypes of ovarian cancer in descending order of frequency are: high-grade serous carcinoma, clear cell carcinoma, endometrioid carcinoma, mucinous carcinoma, and low-grade serous carcinoma, from which only high-grade serous carcinoma shows high response rates to platinum-based chemotherapy.
With more than one in four women not responding to standard first-line chemotherapy treatment, having alternate therapies available is critical. By more accurately identifying and assigning ovarian cancers to their proper histotype, there is greater opportunity to personalize treatment and potentially improve outcomes for these patients. Histotype- specific genetic counselling is also emerging, investigating the family risk for certain histotypes (e.g. Lynch syndrome for endometrioid).
The present study was published in the International Journal of Gynecological Pathology (September 2016) http://journals.lww.com/intjgynpathology/Fulltext/2016/09000/An_Immunohistochemical_Algorithm_for_Ovarian.6.aspx
Using pooled samples including over 1,000 samples from COEUR, the researchers reclassified histotype using immunohistochemical (IHC) biomarkers. The validity of reclassification was supported by gold standard microscopical review, clinical outcome differences and mutational patterns detected by targeted sequencing in a subset of cases. The highest misclassification occurred in the endometrioid histotype, where most of the changes involved reclassification from endometrioid to high-grade serous carcinoma (the most lethal form).
Then 1,762 reclassified cases were subjected to several statistical prediction models with a variable IHC marker input. The agreement of the statistical prediction with four IHC markers as input (i.e. WT1, TP53, NAPSA and PGR) was 88% and increased to 93% using an eight marker input. In other words, a limited IHC marker panel can correctly sub-classify approximately 90% of ovarian carcinomas. In conjunction with gold-standard microscopic evaluation, which also has about 90% accuracy, histotype can be assigned with greater than 95% accuracy.
When treatment options were limited, distinguishing between the different subtypes of ovarian cancer was not as important clinically. Yet, with a growing number of targeted and novel therapies, it is becoming increasingly important to match the patient's cancer with the most appropriate treatment – a goal that starts with a specific and accurate diagnosis. While this immunohistochemical tool can be used for retrospective research cohorts, it can also be applied for clinical trials inclusion, which will likely become histotype-specific disease in their design.
Study: An immunohistochemical algorithm for ovarian carcinoma typing
Authors: Martin Köbel, Kurosh Rahimi, Peter F. Rambau, Christopher Naugler, Cécile Le Page, Liliane Meunier, Manon de Ladurantaye, Sandra Lee, Samuel Leung, Ellen L. Goode, Susan J. Ramus, Joseph W. Carlson, Xiaodong Li, Carol A. Ewanowich, Linda E. Kelemen, Barbara Vanderhyden, Diane Provencher, David Huntsman, Cheng-Han Lee, C. Blake Gilks, and Anne-Marie Mes Masson
Funding: The specimen collection for the COEUR cohort was supported by a grant from the Terry Fox Research Institute. Clinical specimens from the province of Quebec were provided by the Banque de tissus et de données of the Réseau de recherche sur le cancer of the Fonds de recherche du Québec-Santé, which is affiliated to the Canadian Tumor Repository Network. Biologic materials from the province of Ontario were provided by the Ottawa Ovarian Cancer Tissue Bank, the University Health Network Biobank and Ontario Tumour Bank,
the later being funded by the Ontario Institute for Cancer Research. Specimens from the province of British Columbia were provided by the BCCA Tumor Tissue Repository and the OvCare Gynecologic Tissue Bank. Finally, specimens from the province of Alberta were provided from the CBCF Tumor Biobank. The AOVT study was supported by the Canadian Institutes of Health Research (MOP-86727).
Links#2